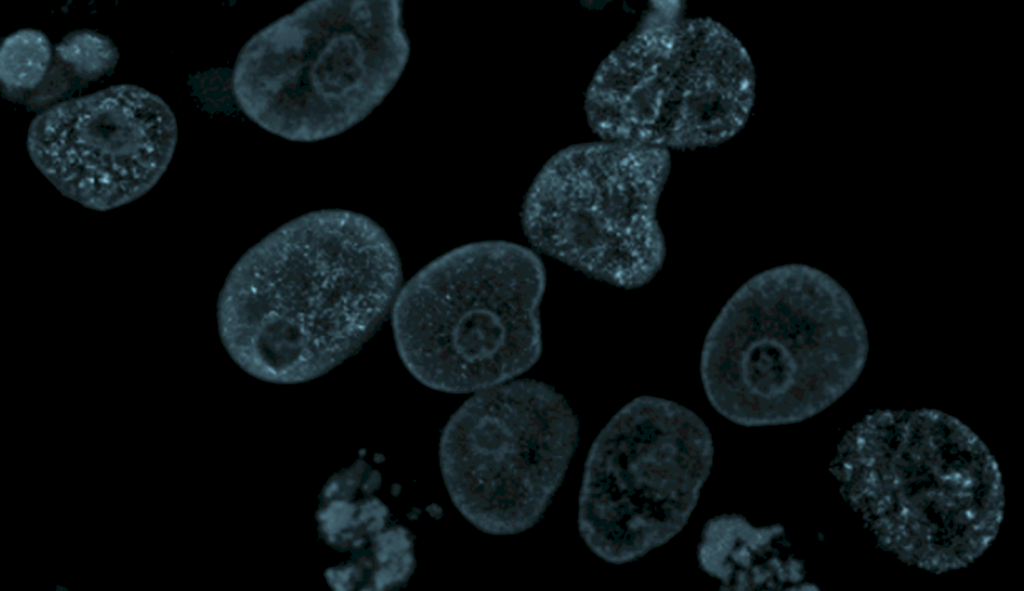
Single Cell Isolation Methods
The first step in single cell sequencing is isolating individual cells from a heterogeneous population. Various techniques have been developed to ensure efficient and gentle isolation, preserving cell viability and integrity.
- Microfluidics-based Techniques: Microfluidics involves manipulating small volumes of fluids on a microscale, allowing precise control over cell trapping and separation. Platforms like droplet-based microfluidics and micro-well arrays encapsulate individual cells in nanoliter-sized droplets or microwells, enabling high-throughput single cell isolation.
- Laser Capture Microdissection (LCM): LCM utilizes laser microdissection to isolate specific cells or regions of interest from tissue samples. It preserves the spatial context of the cells within the tissue, making it invaluable for studying complex tissues and heterogeneous samples.
- Fluorescence-Activated Cell Sorting (FACS): FACS is a flow cytometry-based technique that isolates cells based on their physical and fluorescent properties. Labeled with fluorescent markers, cells are sorted based on their fluorescence intensity, offering high-speed and high-purity cell sorting.
Single Cell DNA Sequencing
Single cell DNA sequencing explores the genetic landscape of individual cells, providing insights into genomic variations, mutations, and structural changes.
- Whole Genome Amplification (WGA) Techniques: WGA is critical in single cell DNA sequencing, amplifying the entire genome from a single cell. Techniques like multiple displacement amplification (MDA) and multiple annealing and looping-based amplification cycles (MALBAC) overcome the limited DNA available in a single cell, enabling downstream sequencing and analysis.
- Single Nucleotide Polymorphism (SNP) Analysis: SNPs are variations in a single nucleotide in the genome. Single cell SNP analysis provides insights into genetic diversity, population structure, and disease-associated variants, enabling the identification of rare variants and somatic mutations.
- Copy Number Variation (CNV) Analysis: CNVs involve gains or losses of DNA segments in the genome. Single cell CNV analysis reveals genomic instability, clonal evolution, and the impact of CNVs on disease development.
Single Cell RNA Sequencing (scRNA-seq)
Understanding gene expression at the single-cell level is essential for unraveling cellular diversity and dynamics.
- mRNA Sequencing (mRNA-Seq): mRNA-Seq captures the transcriptome of individual cells, allowing the quantification of gene expression, identification of cell types, and discovery of novel transcripts.
- Single Cell Expression Profiling: These techniques focus on capturing specific gene subsets in individual cells, enabling targeted analysis of gene families or cellular functions.
- Single Cell Transcriptome Assembly: This computational approach reconstructs full-length transcripts from short sequencing reads, identifying alternative splicing events and isoform diversity.
Single Cell Epigenetic Sequencing
Single cell epigenetic sequencing investigates heritable changes in gene expression patterns that do not involve alterations in the DNA sequence.
- DNA Methylation Analysis: This method assesses DNA methylation patterns in individual cells, providing insights into cell identity, differentiation, and disease progression.
- Chromatin Accessibility Profiling: Techniques like ATAC-seq measure the accessibility of DNA within the chromatin structure, reflecting the regulatory potential of specific genomic regions.
- Histone Modification Analysis: This analysis investigates specific histone marks in individual cells, providing insights into gene regulation and cellular states.
Data Analysis in Single Cell Sequencing
Single cell sequencing generates vast amounts of data, requiring robust computational analysis methods to extract meaningful insights.
Preprocessing and Quality Control
Preprocessing steps, including quality control, read alignment, and removal of technical artifacts, ensure data quality and reliability.
Dimensionality Reduction Techniques
Techniques like Principal Component Analysis (PCA) and t-Distributed Stochastic Neighbor Embedding (t-SNE) transform high-dimensional data into a lower-dimensional space, enabling visualization and interpretation.
Clustering and Cell Type Identification
Clustering algorithms group cells with similar gene expression profiles, while computational approaches assign cell types to clusters, providing insights into cell population diversity.
Trajectory Analysis and Pseudotime Estimation
These methods reconstruct developmental trajectories and infer the order of cellular states, providing insights into cellular transitions and lineage relationships.
Ethical Considerations and Privacy Issues
As with any technology involving large-scale genomic data, single cell sequencing raises ethical considerations and privacy issues. Strict data privacy protocols, informed consent, and ethical guidelines are crucial to ensuring the responsible use of single cell sequencing data.
Future Directions in Single Cell Sequencing
The field of single cell sequencing is rapidly evolving, with emerging technologies like spatial transcriptomics and single-cell proteomics expanding the possibilities of single cell analysis. As technology advances, single cell sequencing techniques are expected to become more scalable, sensitive, and cost-effective, broadening their accessibility and application.