What is RNA-seq?
RNA-sequencing (RNA-seq) is a powerful technique that analyzes the quantity and sequences of RNA in a sample using next-generation sequencing (NGS). By examining the transcriptome, RNA-seq reveals which genes encoded in our DNA are active (turned on or off) and in what quantities they are expressed.
Why is RNA-seq Used?
RNA-seq leverages high-throughput sequencing technologies to provide deep insights into a cell’s transcriptome. Compared to older methods like Sanger sequencing and microarray-based techniques, RNA-seq offers much higher coverage and greater resolution, enabling a more detailed and accurate view of the transcriptome’s dynamic nature.
Applications of RNA-seq
RNA-seq enables the exploration of the entire transcriptome, which includes all types of RNA such as mRNA, rRNA, and tRNA. Understanding the transcriptome is crucial for linking genomic information to functional protein expression. RNA-seq provides key insights into gene activity, transcription levels, and the timing of gene expression, which are essential for understanding cellular biology and identifying potential disease indicators.
Popular applications of RNA-seq include:
- Transcriptional Profiling: Identifying which genes are active in a cell and quantifying their expression levels.
- SNP Detection: Discovering single nucleotide polymorphisms that may impact gene function.
- RNA Editing: Analyzing post-transcriptional modifications in RNA sequences.
- Differential Gene Expression Analysis: Comparing gene expression across different conditions or treatments to identify significant changes.
RNA-seq also captures data on alternative splicing events, producing multiple transcripts from a single gene sequence, and post-transcriptional modifications during mRNA processing, both of which are missed by traditional DNA sequencing methods.
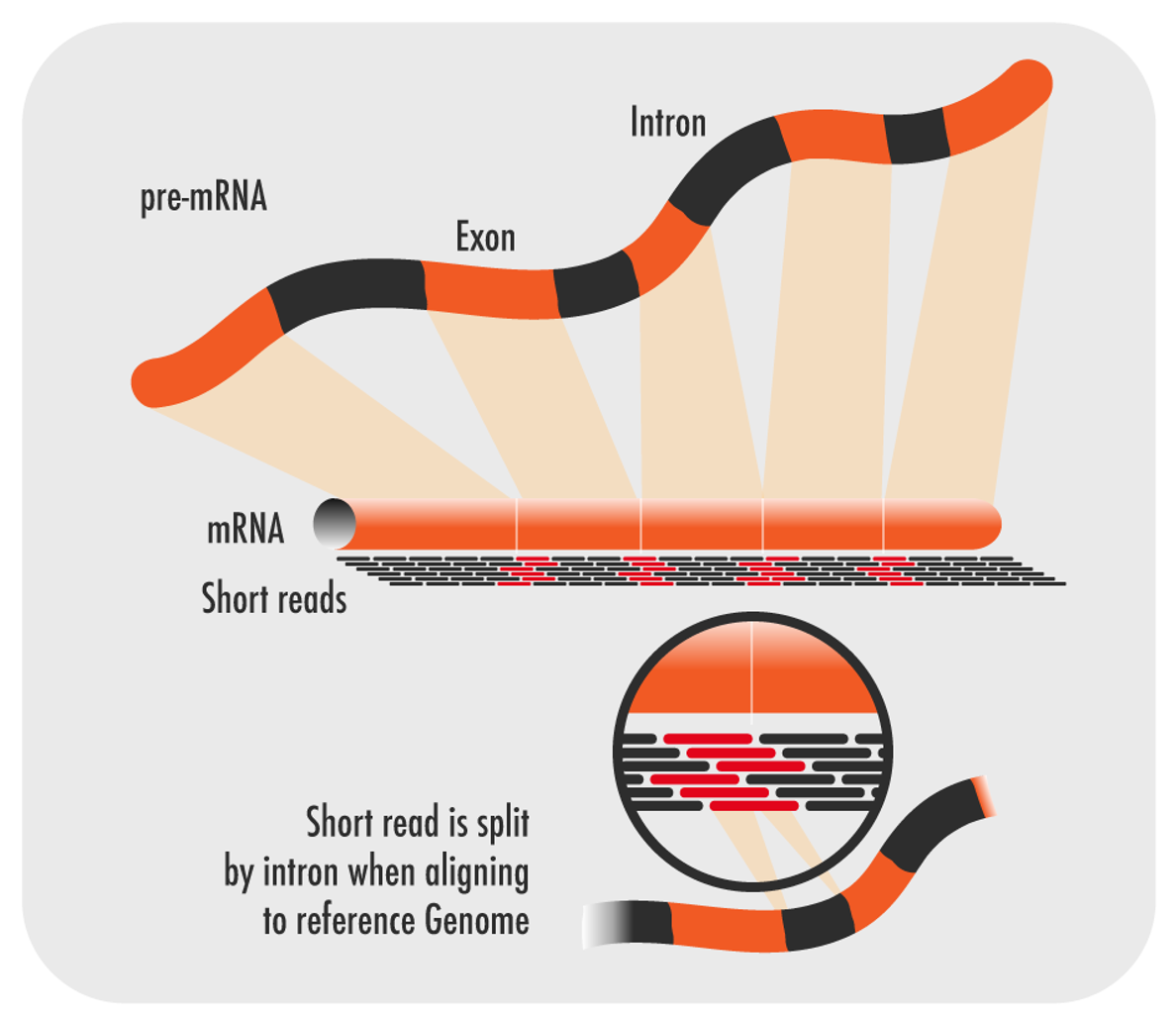
Figure 2: Diverse Applications of RNA-seq in Modern Genomics
RNA-seq Methods
Several RNA-seq methods are tailored to specific research needs:
- mRNA Sequencing: Precisely assesses gene expression, identifies known and novel isoforms, detects gene fusions, and measures allele-specific expression.
- Targeted RNA Sequencing: Focuses on the expression of specific genes of interest, using either enrichment or amplicon-based approaches.
- Ultra-Low-Input and Single-Cell RNA-seq: Ideal for studying processes like differentiation, proliferation, and tumorigenesis at the single-cell level.
- RNA Exome Capture Sequencing: Targets the coding regions of the transcriptome, making it suitable for low-quality samples or limited material.
- Total RNA Sequencing: Measures gene and transcript abundance across coding and noncoding RNA.
- Small RNA Sequencing: Isolates and sequences small RNA species, such as microRNAs, to understand their role in gene regulation.
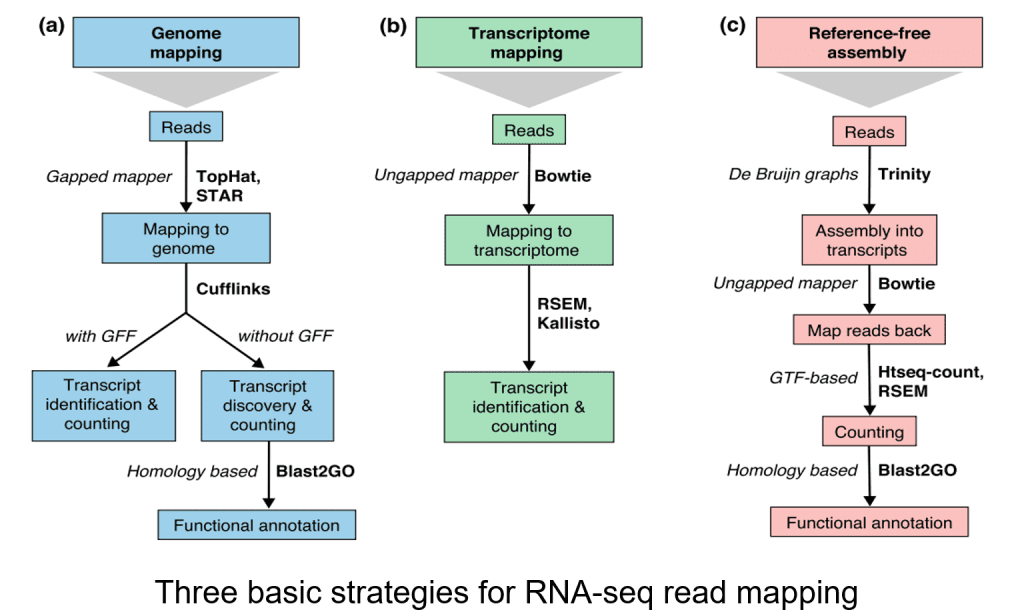
Bioinformatics Workflow of RNA-seq
RNA-seq data analysis involves multiple steps, each crucial for accurate interpretation:
Step 1: Quality Control of Raw Reads
Quality control ensures the accuracy of RNA-seq data by analyzing sequencing quality, GC content, adapter content, overrepresented k-mers, and duplicated reads. Low-quality bases should be trimmed to improve read alignment.
Step 2: Read Alignment
Reads are aligned to a reference genome or transcriptome, or assembled de novo if no reference is available. Accurate alignment is essential for identifying gene isoforms and other transcriptional variations.
Step 3: Transcript Quantification
This step estimates gene and transcript expression levels, providing a quantitative measure of RNA-seq data.
Differential Expression Testing
This process evaluates whether a gene is differentially expressed between conditions. Various normalization techniques are applied before comparison to ensure accurate results.
Alternative Splicing Analysis
Alternative splicing generates multiple transcripts from a single gene, adding complexity to gene expression. Specialized bioinformatics tools are used to detect and analyze these splicing events.
Visualization
Visualization tools, such as genome browsers and RNA-seq specific software, are essential for interpreting RNA-seq data. These tools help researchers explore gene expression, alternative splicing, and other features of the transcriptome.
Functional Profiling
The final step in RNA-seq analysis is functional profiling, which involves identifying the molecular functions and pathways associated with differentially expressed genes. Tools like Gene Ontology, DAVID, and Blast2GO are commonly used for this purpose.
Advanced RNA-seq Analysis
Advanced analysis often involves integrating RNA-seq data with other technologies to gain deeper insights. For more information on the applications of RNA-seq, explore our comprehensive resources.
Benefits of RNA Sequencing
- Broad Dynamic Range: Enables sensitive and accurate measurement of gene expression.
- Unbiased Discovery: Captures both known and novel features without prior knowledge.
- Species Flexibility: Applicable to any species, even those without reference sequences.
- Cost-Effective: Often provides better value compared to traditional methods, with comparable or lower costs per sample.