Introduction
With the advent of the 21st century, we’ve witnessed remarkable technological advancements, particularly in the field of high-throughput ‘omics’ techniques such as microarray analysis and next-generation sequencing. These cutting-edge techniques have revolutionized biological research by generating vast amounts of data, enabling the simultaneous measurement or sequencing of thousands of genes. By conducting a single experiment, researchers can now identify associations between genes and phenotypes. However, the challenge lies in accurately interpreting this complex data.
To address this challenge, numerous computational tools and programs have been developed, enabling the categorization of gene expression profiles into more manageable functional categories. This process, known as Functional Enrichment Analysis (FEA) or Gene Set Analysis (GSA), plays a crucial role in uncovering biological annotations that are over-represented in a gene list compared to a reference background. By doing so, FEA helps researchers understand the biological processes and molecular mechanisms associated with specific experimental conditions.
Types of Functional Enrichment Analysis
Functional Enrichment Analysis, or GSA, can be classified into three primary types:
- Singular Enrichment Analysis (SEA)
- Gene Set Enrichment Analysis (GSEA)
- Modular Enrichment Analysis (MEA)
Singular Enrichment Analysis (SEA)
Singular Enrichment Analysis (SEA) is a conventional approach that determines the statistical significance of individual annotations—such as functional or pathway terms—within a candidate gene list, such as differentially expressed genes. In SEA, preselected genes of interest are analyzed to test the enrichment of each annotation term one by one. The results are typically presented in a tabular format, ordered by the enrichment probability (P-value).
Statistical methods like the Chi-square test, Fisher’s exact test, hypergeometric distribution, and binomial probability are employed to calculate the enriched P-values. SEA is an efficient method for interpreting large gene lists generated by high-throughput technologies.
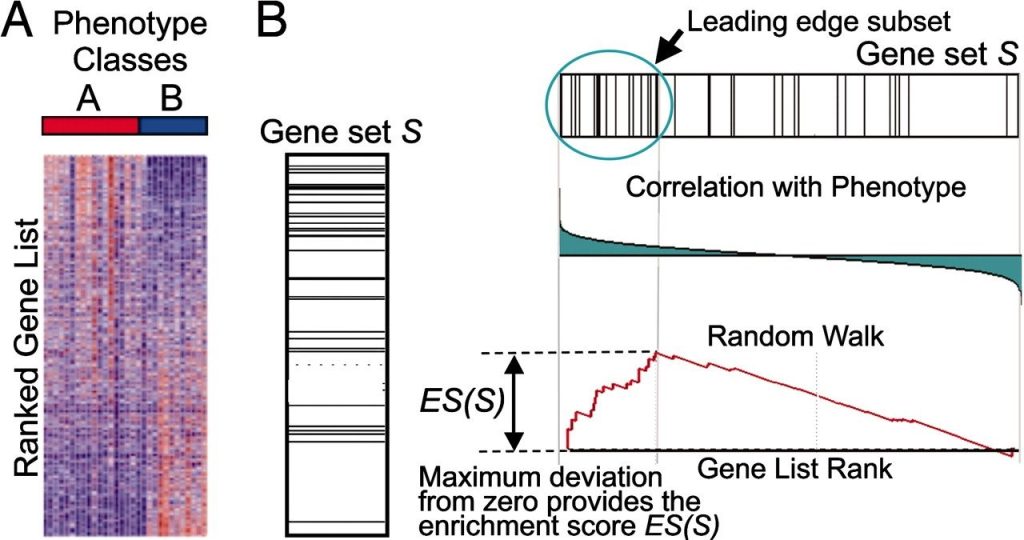
Gene Set Enrichment Analysis (GSEA)
Gene Set Enrichment Analysis (GSEA) represents a more advanced type of GSA. It evaluates the distribution of genes linked to a given term across the entire experimental dataset, with genes ranked according to specific criteria, such as fold change. GSEA is particularly useful for identifying gene sets with consistent differences between two biological states, such as distinct phenotypes.
A typical GSEA workflow involves:
- Enrichment Score (ES) Calculation: This step measures the degree of over-representation of a gene set at the extremes of a ranked gene list.
- Significance Level Estimation: The statistical significance of the ES is determined through a permutation test that accounts for the complex structure of gene expression data.
- Adjustment for Multiple Hypothesis Testing: The ES is normalized to account for gene set size, and the false discovery rate (FDR) is calculated to control for false positives.
Modular Enrichment Analysis (MEA)
Modular Enrichment Analysis (MEA) takes FEA a step further by considering the relationships between annotation terms. By integrating network discovery algorithms with basic enrichment calculations from SEA, MEA uncovers unique biological meanings that cannot be deduced from isolated terms. This approach is particularly valuable when dealing with heterogeneous annotation content, as it reduces redundancy and highlights interrelated biological processes.
Categorization of Functional Enrichment Tools
Functional enrichment tools can be broadly categorized into two main types:
- Over-Representation Analysis (ORA)
- Functional Class Scoring (FCS)
Over-Representation Analysis (ORA)
ORA is an extension of single-gene analysis and is one of the most widely used approaches in gene set analysis due to its simplicity and well-understood statistical model. ORA involves querying differentially expressed genes (DEGs) against curated pathways and performing statistical tests to determine whether the number of DEGs associated with a specific gene set exceeds random chance.
However, when using ORA for differential gene expression analysis, it is essential to select an appropriate background gene list. Using the entire genome as a background may not be suitable, especially in tissue-specific studies where many genes are not expressed.
Functional Class Scoring (FCS)
Unlike ORA, Functional Class Scoring (FCS) methods utilize the entire expression matrix rather than just a list of DEGs. This approach addresses the limitations of ORA by considering the collective impact of genes within a biological process, recognizing that genes and proteins often act in groups. FCS methods can be further divided into univariate and multivariate approaches, depending on how the gene set scores are calculated.
Topology-Based Pathway Analysis
In topology-based pathway analysis, not all genes within a pathway are considered equal. The inclusion of pathway topology information, such as gene product interactions, enhances the accuracy of enrichment analysis by quantifying the importance of specific genes within a pathway.
Essential Databases for Functional Enrichment Analysis
Two critical databases are central to functional enrichment analysis:
- Gene Ontology (GO)
- Kyoto Encyclopedia of Genes and Genomes (KEGG)
Gene Ontology (GO)
Gene Ontology (GO) is the cornerstone of FEA, enabling knowledge-based computational analysis of biological data from large-scale assays. GO organizes our understanding of biological systems into three main aspects: molecular function, cellular component, and biological process. It provides a hierarchical structure that helps researchers identify over-represented functions within a gene set.
Kyoto Encyclopedia of Genes and Genomes (KEGG)
KEGG is a comprehensive database resource that integrates genomic and chemical information with knowledge of molecular wiring diagrams. It allows researchers to explore high-level functions and biological significance across various systems, from cellular processes to entire ecosystems.
Conclusion
Functional Enrichment Analysis is indispensable in analyzing the vast amounts of data generated by high-throughput techniques. It enables researchers to assign functions to previously unknown genes and uncover crucial biological pathways. While numerous tools are available for FEA, each has its limitations, underscoring the ongoing need for a gold-standard tool in functional enrichment analysis.